Structures
0 Human milk oligosaccharides match composition H3
Show / hide: structures
Jump to: statistics | enzymes
Show / hide: structures
Jump to: statistics | enzymes
Click on a glycan to open it in GlycoForm
| Download structures as: | Hydrolyse these structures: go |
Glycan profile
Lex: 0 (n/a) SLex: 0 (n/a) Lea: 0 (n/a) SLea: 0 (n/a) Leb: 0 (n/a) Ley: 0 (n/a) A: 0 (n/a) B: 0 (n/a) H: 0 (n/a) Sda/Cad: 0 (n/a)
Enzyme profile
0 enzymes are knocked out Change| Knockout: | |
Reaction network
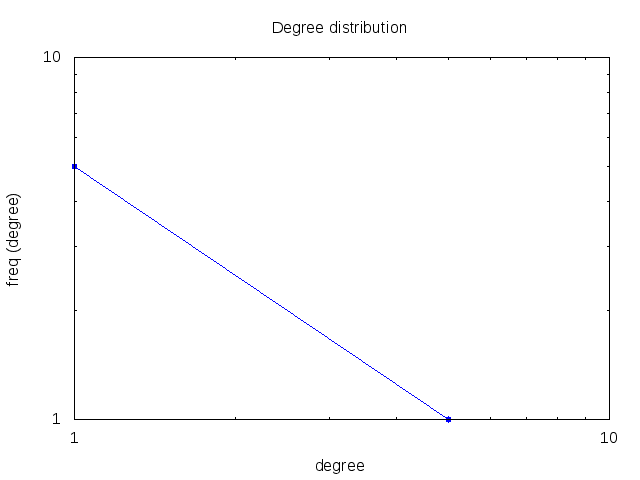
| |||||
|
Colour key: N-acetylgalactosaminyltransferases (brown); N-acetylglucosaminyltransferases (blue); fucosyltransferases (red); galactosyltransferases (yellow); sialyltransferases (magenta); sulfotransferases (orange)